Overview¶
A high-performance topological machine learning toolbox in Python
giotto-tda is a high performance topological machine learning toolbox in Python built on top of
scikit-learn and is distributed under the GNU AGPLv3 license. It is part of the Giotto family of open-source projects.
Guiding principles¶
scikit-learnStrictly adhere to thescikit-learnAPI and development guidelines, inherit the strengths of that framework.joblib).
30s guide to giotto-tda¶
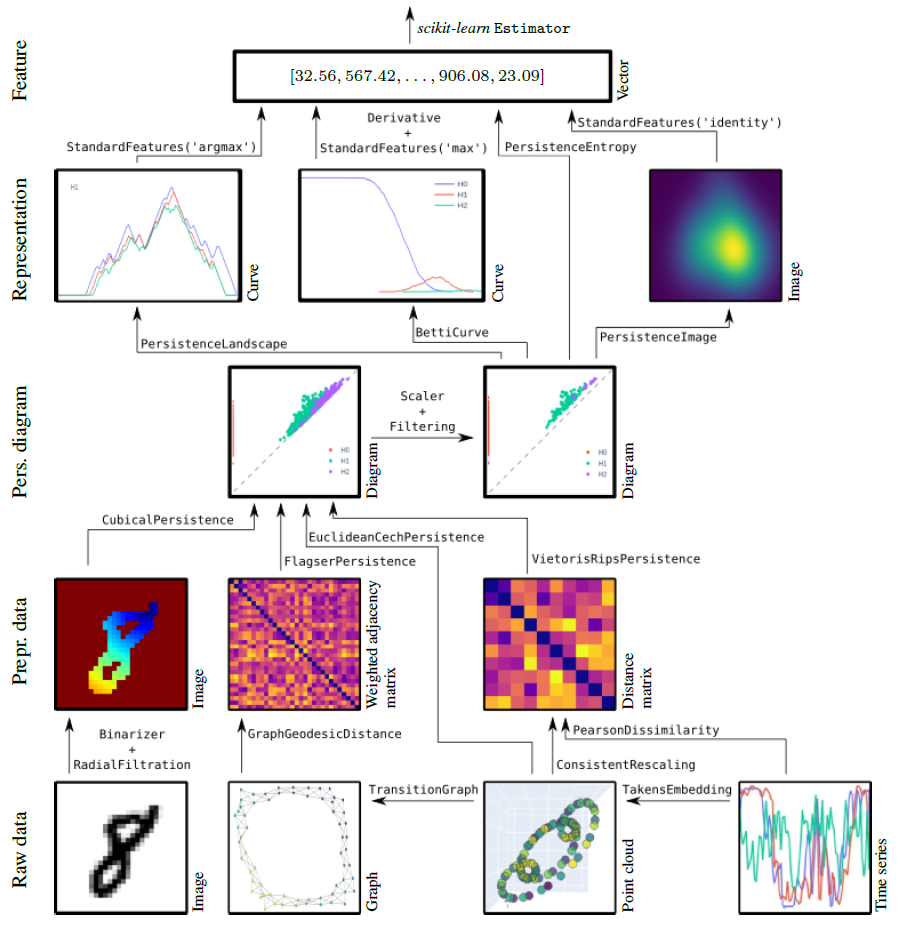
For installation instructions, see the installation instructions.
The functionalities of giotto-tda are provided in scikit-learn–style transformers.
This allows you to generate topological features from your data in a familiar way. Here is an example with the VietorisRipsPersistence transformer:
from gtda.homology import VietorisRipsPersistence
VR = VietorisRipsPersistence()
which computes topological summaries, called persistence diagrams, from collections of point clouds or weighted graphs, as follows:
diagrams = VR.fit_transform(point_clouds)
A plotting API allows for quick visual inspection of the outputs of many of giotto-tda’s transformers. To visualize the i-th output sample, run
diagrams = VR.plot(diagrams, sample=i)
You can create scalar or vector features from persistence diagrams using giotto-tda’s dedicated transformers. Here is an example with the PersistenceEntropy transformer:
from gtda.diagrams import PersistenceEntropy
PE = PersistenceEntropy()
features = PE.fit_transform(diagrams)
features is a two-dimensional numpy array. This is important to making this type of topological feature generation fit into a typical machine learning workflow from scikit-learn.
In particular, topological feature creation steps can be fed to or used alongside models from scikit-learn, creating end-to-end pipelines which can be evaluated in cross-validation,
optimised via grid-searches, etc.:
from sklearn.ensemble import RandomForestClassifier
from gtda.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(point_clouds, labels)
RFC = RandomForestClassifier()
model = make_pipeline(VR, PE, RFC)
model.fit(X_train, y_train)
model.score(X_valid, y_valid)
giotto-tda also implements the Mapper algorithm as a highly customisable scikit-learn Pipeline, and provides simple plotting functions for visualizing output Mapper graphs and have real-time interaction with the pipeline parameters:
from gtda.mapper import make_mapper_pipeline
from sklearn.decomposition import PCA
from sklearn.cluster import DBSCAN
pipe = make_mapper_pipeline(filter_func=PCA(), clusterer=DBSCAN())
plot_interactive_mapper_graph(pipe, data)
Resources¶
Tutorials and examples¶
We provide a number of tutorials and examples, which offer:
quick start guides to the API;
in-depth examples showcasing more of the library’s features;
intuitive explanations of topological techniques.
Use cases¶
A selection of use cases for giotto-tda is collected at this page.
Please note, however, that some of these were written for past versions of giotto-tda. In some cases,
only small modifications are needed to run them on recent versions, while in others it is best to install
the relevant past version of giotto-tda (preferably in a fresh environmnent). In a couple of cases,
the legacy giotto-learn or giotto-learn-nightly will be needed.
What’s new¶
Major Features and Improvements¶
Wheels for Python 3.9 have been added (#528).
Weighted Rips filtrations, and in particular distance-to-measure (DTM) based filtrations, are now supported in
ripserand by the newWeightedRipsPersistencetransformer (#541).See “Backwards-Incompatible Changes” for major improvements to
ParallelClusteringand thereforemake_mapper_pipelinewhich are also major breaking changes.GUDHI’s edge collapser can now be used with arbitrary vertex and edge weights (#558).
GraphGeodesicDistancecan now take rectangular input (the number of vertices is inferred to bemax(x.shape)), andKNeighborsGraphcan now take sparse input (#537).VietorisRipsPersistencenow takes ametric_paramsparameter (#541).
Bug Fixes¶
A documentation bug affecting plots from
DensityFiltrationhas been fixed (#540).A bug affecting the bindings for GUDHI’s edge collapser, which incorrectly did not ignore lower diagonal entries, has been fixed (#538).
Symmetry conflicts in the case of sparse input to
ripserandVietorisRipsPersistenceare now handled in a way true to the documentation, i.e. by favouring upper diagonal entries if different values in transpose positions are also stored (#537).
Backwards-Incompatible Changes¶
The minimum required version of
pyflagseris now 0.4.3 (#537).ParallelClustering.fit_transformnow outputs one array of cluster labels per sample, bringing it closer toscikit-learnconvention for clusterers, and the fitted single clusterers are no longer stored in theclusterers_attribute of the fitted object (#535 and #552).
Thanks to our Contributors¶
This release contains contributions from many people:
Umberto Lupo, Julian Burella Pérez, and Wojciech Reise.
We are also grateful to all who filed issues or helped resolve them, asked and answered questions, and were part of inspiring discussions.